[2]:
import scanpy as sc
from step import scModel, stModel
from step.utils.misc import read_visium_hd
sc.set_figure_params(dpi=150, figsize=(6, 4.5), dpi_save=300)
/projects/82505004-e7a0-445f-ab3c-80d03c91438f/.cache/pypoetry/virtualenvs/step-Ajq_Bw_i-py3.10/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Visium HD Mouse Small Intestine (8 um and 16 um) cell type clustering & spatial domain identification¶
16 um bin size: Cell type clustering¶
[3]:
adata = read_visium_hd("./data/visium-hd/mouse-intesitine/square_016um/")
[4]:
stepc = scModel(
adata=adata,
n_top_genes=2000,
)
Trying seurat_v3 for hvgs
not log_transformed
Adding count data to layer 'counts'
================Dataset Info================
Batch key: None
Class key: None
Number of Batches: 1
Number of Classes: None
Gene Expr: (90987, 2000)
============================================
[5]:
stepc.run(epochs=400, batch_size=1024,)
Performing global random split
Current Mode: multi_batches_with_ct: ['gene_expr', 'class_label', 'batch_label']
66%|██████▌ | 263/400 [18:32<08:58, 3.93s/epoch, kl_loss=0.937, recon_loss=392.133, val_kl_loss=0.933/0.565, val_recon_loss=393.606/392.961]Early Stopping triggered
66%|██████▌ | 263/400 [18:32<09:39, 4.23s/epoch, kl_loss=0.937, recon_loss=392.133, val_kl_loss=0.933/0.565, val_recon_loss=393.606/392.961]
EarlyStopping counter: 30 out of 30
EarlyStopping counter: 263 out of 10
[7]:
adata = stepc.adata
sc.pp.neighbors(adata, use_rep='X_rep', n_neighbors=60)
sc.tl.umap(adata)
[9]:
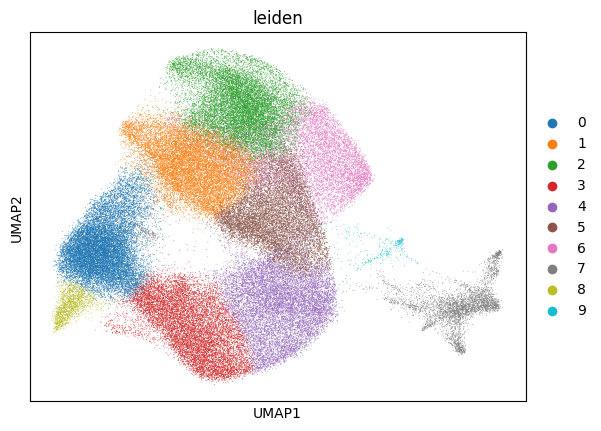
sc.tl.leiden(adata)
sc.pl.umap(adata, color='leiden')
[10]:
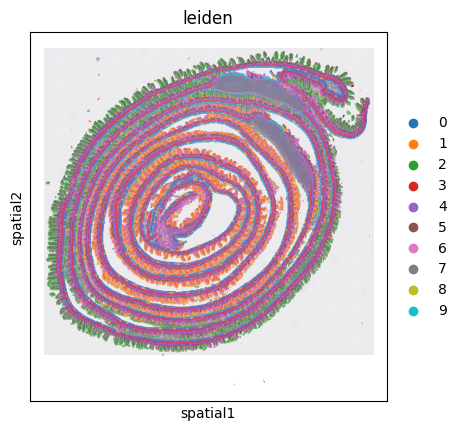
sc.pl.spatial(adata, color='leiden')
[11]:
stepc.save("./results/visium-hd/config")
adata.write_h5ad("./results/visium-hd/mouse_intesitine_16um.h5ad")
Saving model...
Saving model config...
Saving dataset config...
8 um bin size: Spatial domain identification¶
[2]:
adata = read_visium_hd("./data/visium-hd/mouse-intesitine/square_008um/")
[3]:
stepc = stModel(
adata=adata,
n_top_genes=2000,
edge_clip=1,
)
Trying seurat_v3 for hvgs
not log_transformed
Adding count data to layer 'counts'
Dataset Done
================Dataset Info================
Batch key: None
Class key: None
Number of Batches: 1
Number of Classes: None
Gene Expr: (350107, 2000)
============================================
[4]:
stepc.run()
Training with e2e pattern
Training graph with single batch
100%|██████████| 2000/2000 [02:27<00:00, 13.52step/s, recon_loss=287.522, kl_loss=0.118, contrast_loss=0.248, graph_ids=None]
[16]:
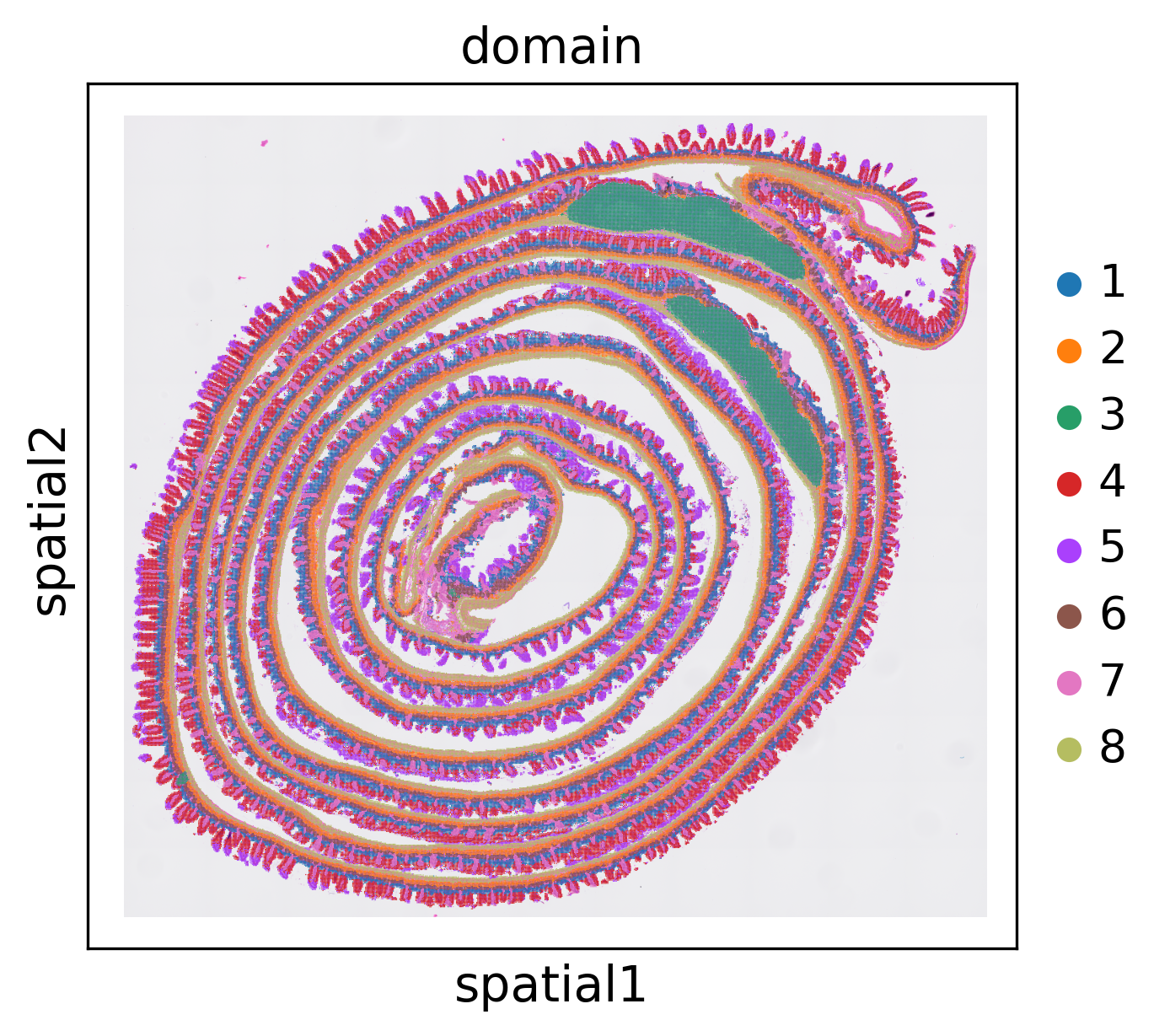
stepc = stModel.load("./results/visium-hd/mouse-intestine/config-8um/",
filepath='./results/visium-hd/mouse_intesitine_8um.h5ad')
stepc.cluster(n_clusters=8)
stepc.spatial_plot(color='domain')
Trying seurat_v3 for hvgs
Adding count data to layer 'counts'
Dataset Done
================Dataset Info================
Batch key: None
Class key: None
Number of Batches: 1
Number of Classes: None
Gene Expr: (326119, 2000)
============================================
Loading backbone model...
Backbone model loaded.
[6]:
import torch
torch.cuda.empty_cache()
stepc.add_embed(key_added='X_rep')
adata = stepc.adata
sc.pp.neighbors(adata, use_rep='X_rep', n_neighbors=60)
sc.tl.umap(adata)
[9]:
stepc.save("./results/visium-hd/mouse-intestine/config-8um")
adata.write_h5ad("./results/visium-hd/mouse_intesitine_8um.h5ad")
Saving model...
Saving model config...
Saving dataset config...
[5]:
adata = sc.read_h5ad("./results/visium-hd/mouse_intesitine_8um.h5ad")
[6]:
sc.pl.umap(adata, color='domain', show=False, save='8um_domain.svg')
WARNING: saving figure to file figures/umap8um_domain.svg
[6]:
<Axes: title={'center': 'domain'}, xlabel='UMAP1', ylabel='UMAP2'>
[21]:
adata = stepc.adata
sc.tl.rank_genes_groups(adata, groupby='domain', method='wilcoxon', dendrogram=False, use_raw=True)
sc.tl.dendrogram(adata, groupby='domain')
WARNING: You’re trying to run this on 2000 dimensions of `.X`, if you really want this, set `use_rep='X'`.
Falling back to preprocessing with `sc.pp.pca` and default params.
[22]:
sc.settings.figdir = "./results/visium-hd/mouse-intestine/config-8um/"
sc.pl.rank_genes_groups_matrixplot(adata,
groupby='domain',
values_to_plot='logfoldchanges',
cmap='RdBu_r',
use_raw=True,
vmin=-4, vmax=4, save="8um_domain.svg")
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/matrixplot_8um_domain.svg
[23]:
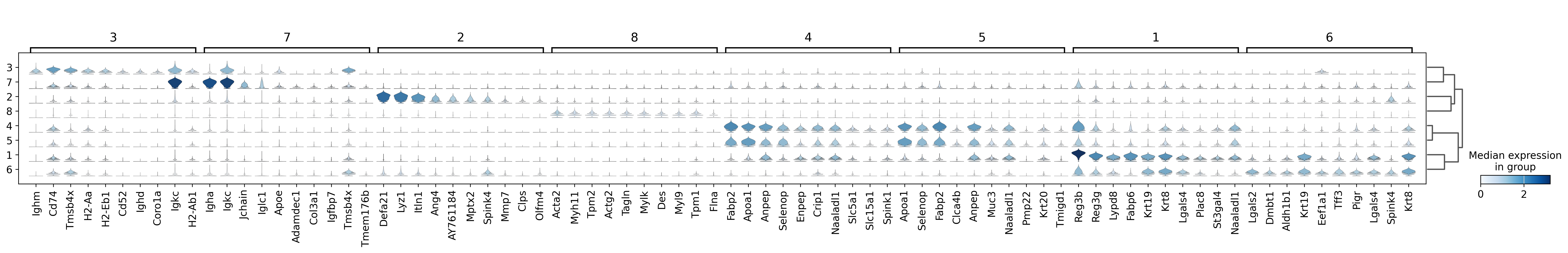
sc.pl.rank_genes_groups_stacked_violin(adata, groupby='domain', use_raw=True, save="8um_domain.svg")
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/stacked_violin_8um_domain.svg
[24]:
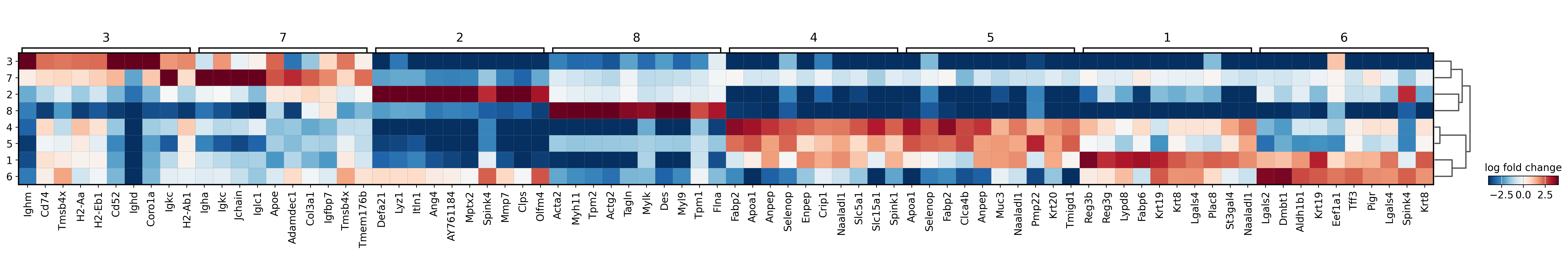
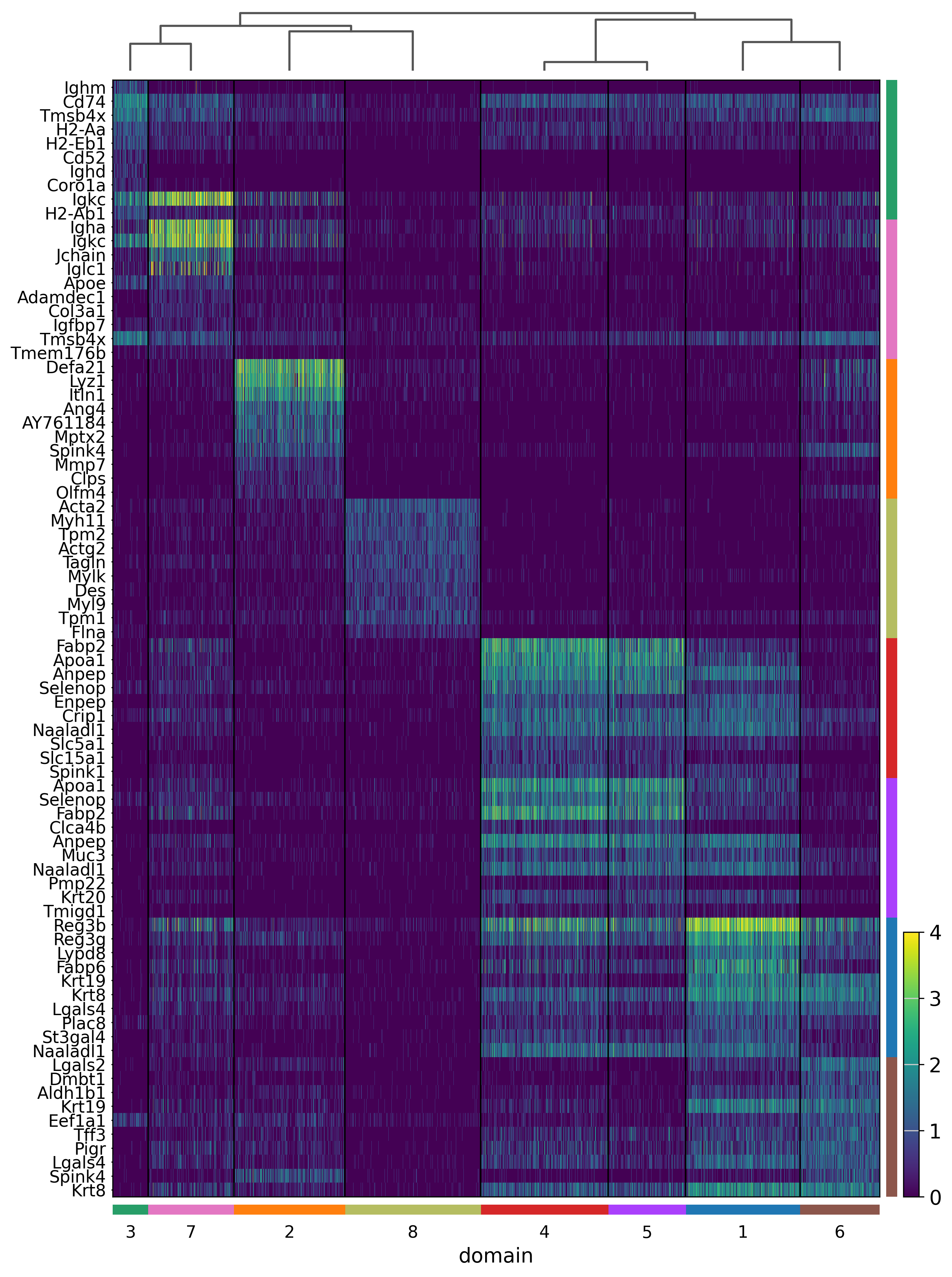
sc.pl.rank_genes_groups_heatmap(adata, groupby='domain', use_raw=True, save="_8um_domain.svg", n_genes=10, swap_axes=True, show_gene_labels=True, vmax=4)
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/heatmap_8um_domain.svg
8 um bin size: Cell type clustering¶
[2]:
adata = read_visium_hd("./data/visium-hd/mouse-intesitine/square_008um/")
[3]:
stepc = scModel(
adata=adata,
n_top_genes=2000,
)
Trying seurat_v3 for hvgs
not log_transformed
Adding count data to layer 'counts'
================Dataset Info================
Batch key: None
Class key: None
Number of Batches: 1
Number of Classes: None
Gene Expr: (350107, 2000)
============================================
[4]:
stepc.run(epochs=400, batch_size=4096)
Performing global random split
Current Mode: single_batch: ['gene_expr']
51%|█████▏ | 205/400 [52:39<1:03:55, 19.67s/epoch, kl_loss=0.870, recon_loss=128.897, val_kl_loss=0.874/0.493, val_recon_loss=127.831/127.675]Early Stopping triggered
51%|█████▏ | 205/400 [52:39<50:05, 15.41s/epoch, kl_loss=0.870, recon_loss=128.897, val_kl_loss=0.874/0.493, val_recon_loss=127.831/127.675]
EarlyStopping counter: 30 out of 30
EarlyStopping counter: 205 out of 10
[5]:
adata = stepc.adata
sc.pp.neighbors(adata, use_rep='X_rep', n_neighbors=60)
sc.tl.umap(adata)
[28]:
sc.tl.leiden(adata, resolution=0.8)
sc.pl.umap(adata, color='leiden')
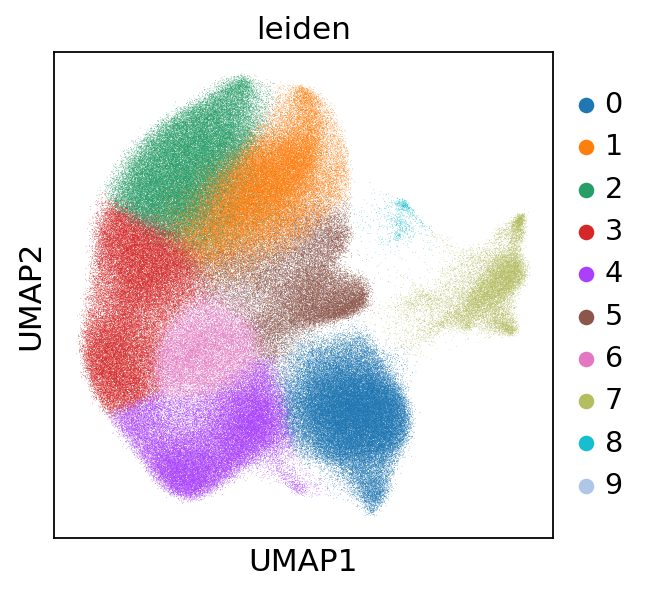
[29]:
adata.obs['leiden'].value_counts()
[29]:
leiden
0 66024
1 59749
2 55412
3 51634
4 48614
5 31971
6 19494
7 15783
8 952
9 474
Name: count, dtype: int64
[30]:
sc.pl.spatial(adata, color='leiden')
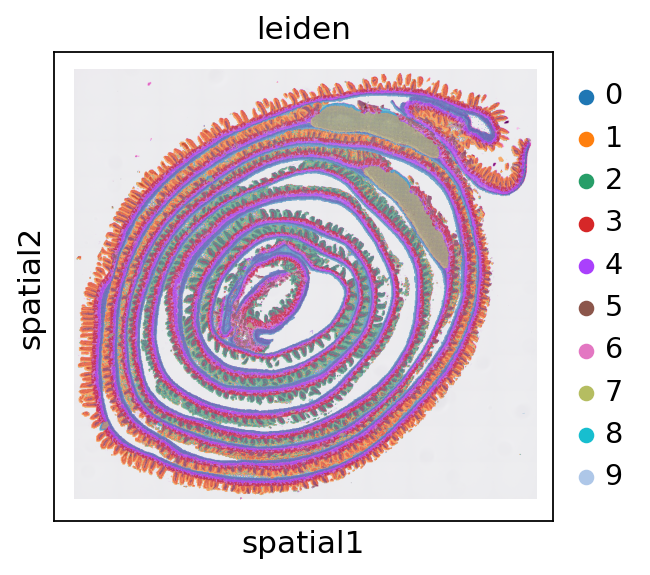
[17]:
# stepc.save("./results/visium-hd/mouse-intestine/config-8um-sc")
adata.write_h5ad("./results/visium-hd/mouse_intesitine_8um_sc.h5ad")
[18]:
# adata = sc.read_h5ad("./results/visium-hd/mouse_intesitine_8um_sc.h5ad")
sc.set_figure_params(dpi_save=300, figsize=(12, 9))
sc.settings.figdir = "./results/visium-hd/mouse-intestine/config-8um/"
sc.pl.spatial(adata, color='leiden', frameon=False, save='_leiden_8um.svg')
sc.pl.umap(adata, color='leiden', frameon=False, save='_leiden_8um.svg', show=False)
# sc.pl.spatial(adata, color='leiden',)
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/show_leiden_8um.svg
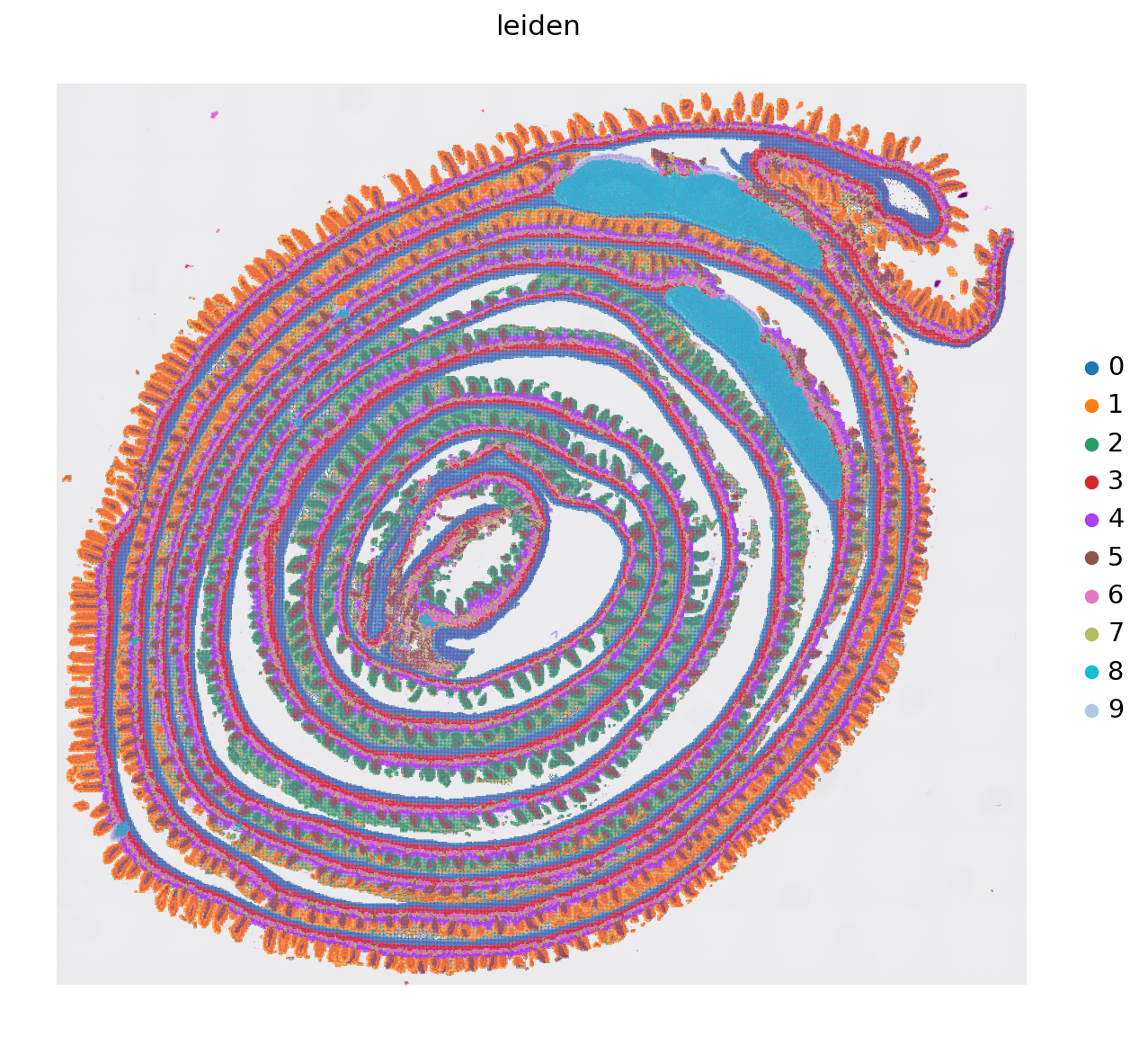
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/umap_leiden_8um.svg
[18]:
<Axes: title={'center': 'leiden'}, xlabel='UMAP1', ylabel='UMAP2'>
[16]:
sc.pl.spatial(adata, color='leiden', groups=['6', '7'], img_key=None)
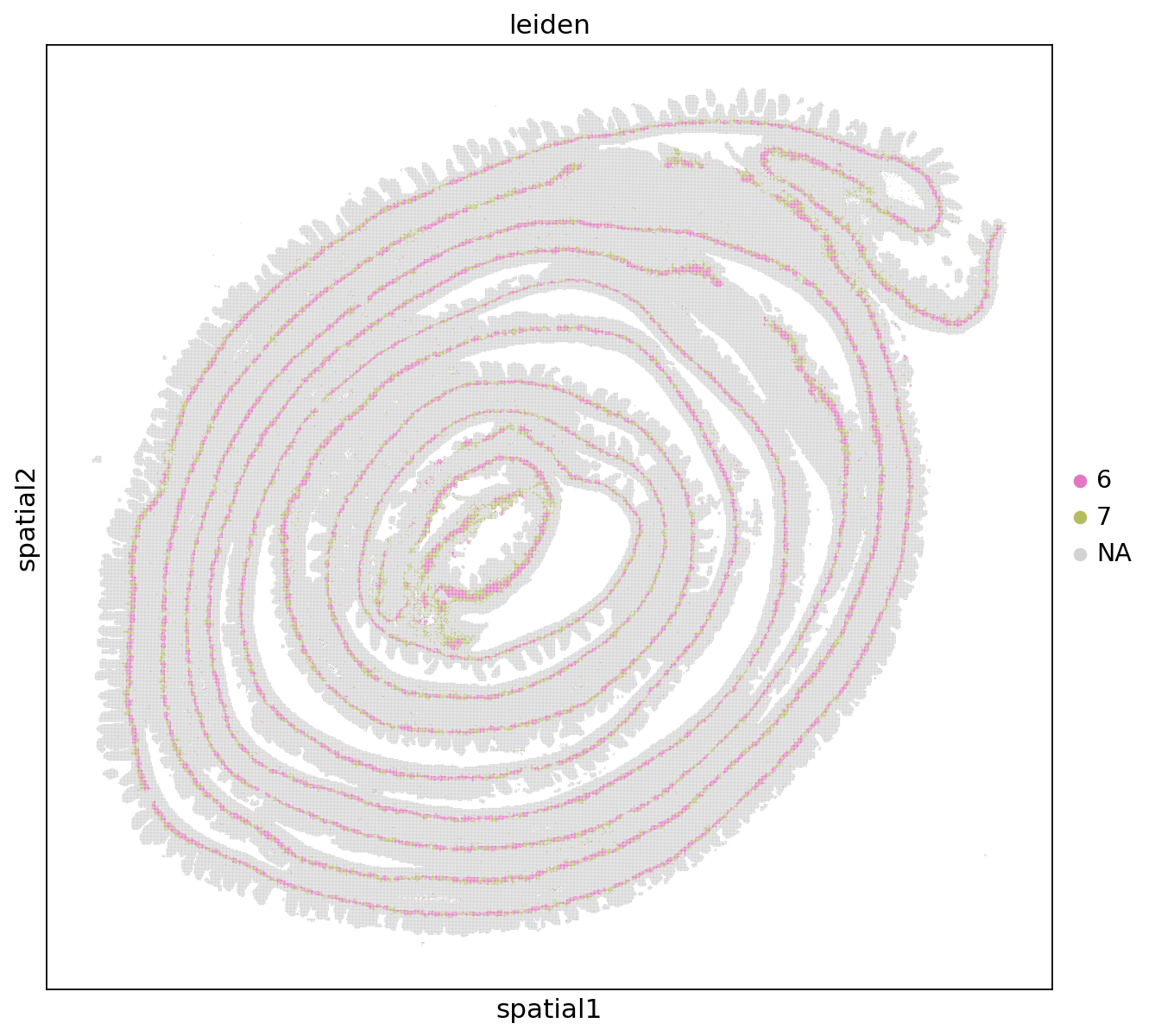
[5]:
adata.obs['leiden'].value_counts()
[5]:
leiden
0 65263
1 56211
2 50772
3 43814
4 38120
5 32429
6 25935
7 19934
8 15772
9 1014
10 836
11 7
Name: count, dtype: int64
[7]:
adata.obs['leiden'] = adata.obs['leiden'].astype(str).apply(lambda x: '0' if x in ['10', '11'] else x)
adata.obs['leiden'] = pd.Categorical(adata.obs['leiden'])
[ ]:
sc.pl.umap(adata, color='leiden', show=False, save='_leiden_8um.svg')
sc.pl.spatial(adata, color='leiden', frameon=False, show=False, save='_leiden_8um.svg')
[10]:
sc.tl.rank_genes_groups(adata, groupby='leiden', method='wilcoxon', dendrogram=False, use_raw=True)
sc.tl.dendrogram(adata, groupby='leiden')
WARNING: You’re trying to run this on 2000 dimensions of `.X`, if you really want this, set `use_rep='X'`.
Falling back to preprocessing with `sc.pp.pca` and default params.
[11]:
sc.settings.figdir = "./results/visium-hd/mouse-intestine/config-8um/"
sc.pl.rank_genes_groups_matrixplot(adata,
groupby='leiden',
values_to_plot='logfoldchanges',
cmap='RdBu_r',
use_raw=True,
vmin=-4, vmax=4, save="8um_ct.svg")
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/matrixplot_8um_ct.svg
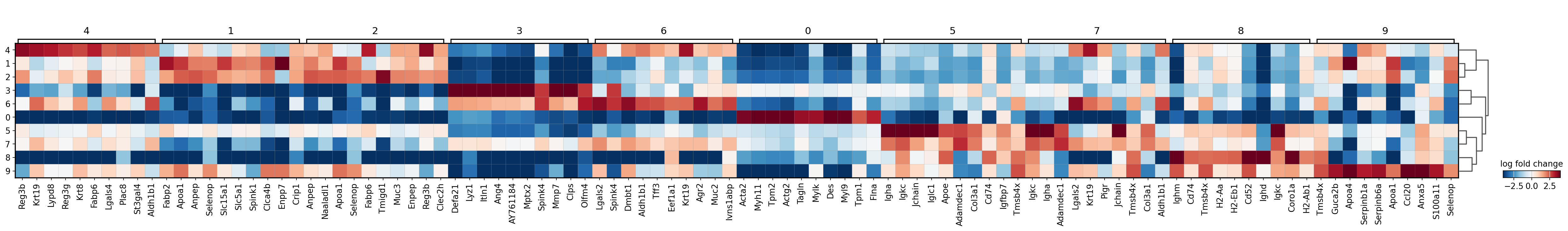
[12]:
sc.pl.rank_genes_groups_stacked_violin(adata, groupby='leiden', use_raw=True, save="8um_ct.svg")
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/stacked_violin_8um_ct.svg
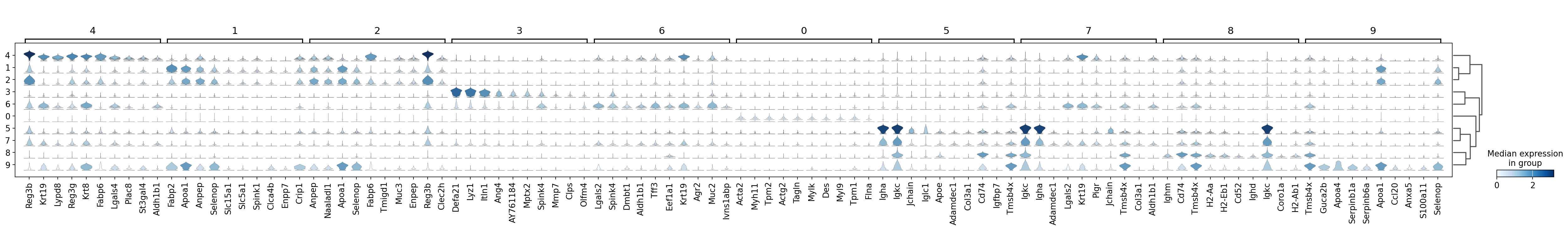
[13]:
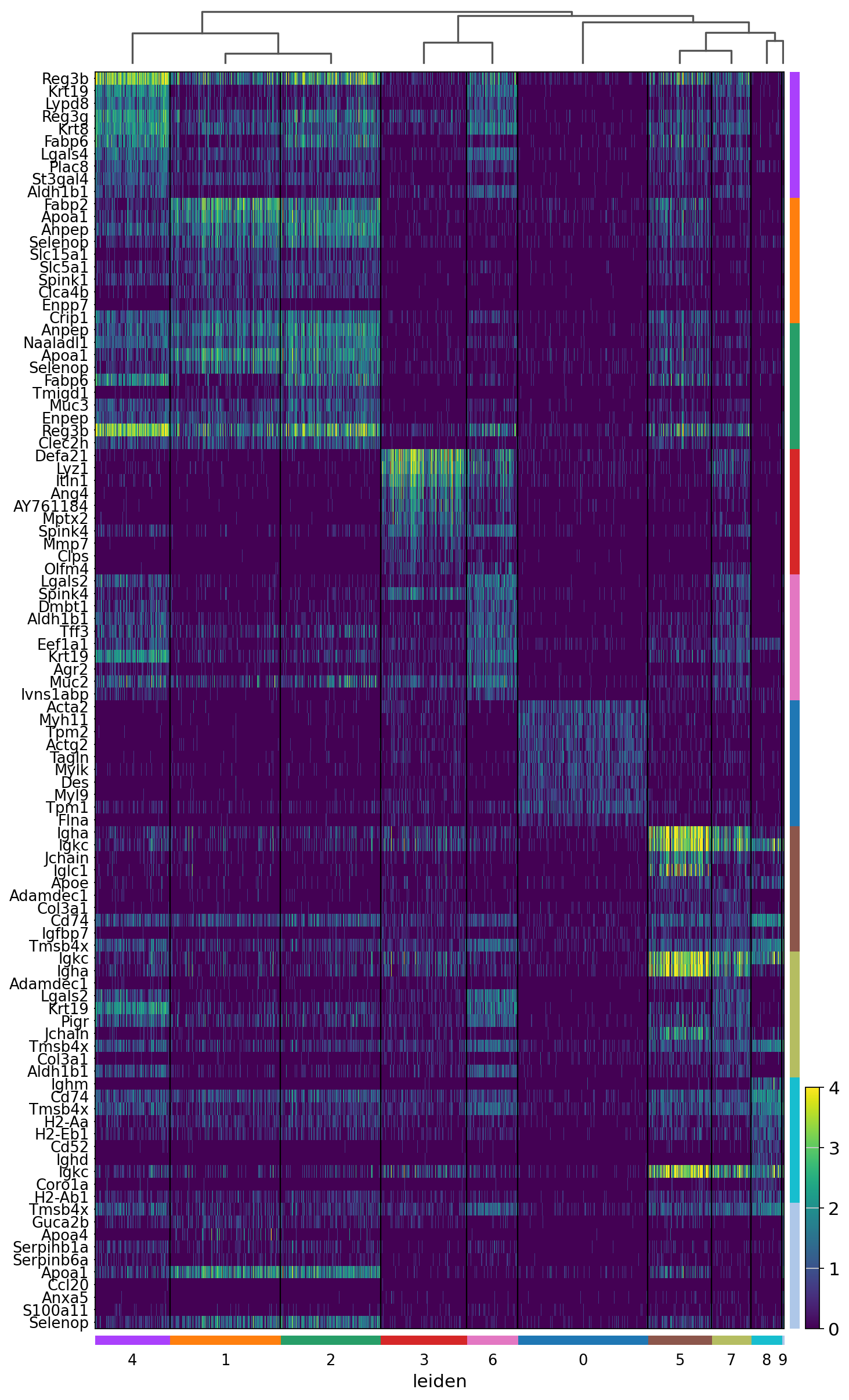
sc.pl.rank_genes_groups_heatmap(adata, groupby='leiden', use_raw=True, save="_8um_ct.svg", n_genes=10, swap_axes=True, show_gene_labels=True, vmax=4)
WARNING: saving figure to file results/visium-hd/mouse-intestine/config-8um/heatmap_8um_ct.svg
[ ]: